About PyMC3¶
Purpose¶
PyMC3 is a probabilistic programming package for Python that allows users to fit Bayesian models using a variety of numerical methods, most notably Markov chain Monte Carlo (MCMC) and variational inference (VI). Its flexibility and extensibility make it applicable to a large suite of problems. Along with core model specification and fitting functionality, PyMC3 includes functionality for summarizing output and for model diagnostics.
Features¶
PyMC3 strives to make Bayesian modeling as simple and painless as possible, allowing users to focus on their scientific problem, rather than on the methods used to solve it. Here is a partial list of its features:
Modern methods for fitting Bayesian models, including MCMC and VI.
Includes a large suite of well-documented statistical distributions.
Uses Theano as the computational backend, allowing for fast expression evaluation, automatic gradient calculation, and GPU computing.
Built-in support for Gaussian process modeling.
Model summarization and plotting.
Model checking and convergence detection.
Extensible: easily incorporates custom step methods and unusual probability distributions.
Bayesian models can be embedded in larger programs, and results can be analyzed with the full power of Python.
What’s new in version 3¶
The third major version of PyMC has benefitted from being re-written from scratch. Substantial improvements in the user interface and performance have resulted from this. While PyMC2 relied on Fortran extensions (via f2py) for most of the computational heavy-lifting, PyMC3 leverages Theano, a library from the Montréal Institute for Learning Algorithms (MILA), for array-based expression evaluation, to perform its computation. What this provides, above all else, is fast automatic differentiation, which is at the heart of the gradient-based sampling and optimization methods currently providing inference for probabilistic programming.
Major changes from previous versions:
New flexible object model and syntax (not backward-compatible with PyMC2).
Gradient-based MCMC methods, including Hamiltonian Monte Carlo (HMC), the No U-turn Sampler (NUTS), and Stein Variational Gradient Descent.
Variational inference methods, including automatic differentiation variational inference (ADVI) and operator variational inference (OPVI).
An interface for easy formula-based specification of generalized linear models (GLM).
Elliptical slice sampling.
Specialized distributions for representing time series.
A library of Jupyter notebooks that provide case studies and fully developed usage examples.
Much more!
While the addition of Theano adds a level of complexity to the development of PyMC, fundamentally altering how the underlying computation is performed, we have worked hard to maintain the elegant simplicity of the original PyMC model specification syntax.
History¶
PyMC began development in 2003, as an effort to generalize the process of building Metropolis-Hastings samplers, with an aim to making Markov chain Monte Carlo (MCMC) more accessible to applied scientists. The choice to develop PyMC as a python module, rather than a standalone application, allowed the use of MCMC methods in a larger modeling framework. By 2005, PyMC was reliable enough for version 1.0 to be released to the public. A small group of regular users, most associated with the University of Georgia, provided much of the feedback necessary for the refinement of PyMC to a usable state.
In 2006, David Huard and Anand Patil joined Chris Fonnesbeck on the development team for PyMC 2.0. This iteration of the software strives for more flexibility, better performance and a better end-user experience than any previous version of PyMC. PyMC 2.2 was released in April 2012. It contained numerous bugfixes and optimizations, as well as a few new features, including improved output plotting, csv table output, improved imputation syntax, and posterior predictive check plots. PyMC 2.3 was released on October 31, 2013. It included Python 3 compatibility, improved summary plots, and some important bug fixes.
In 2011, John Salvatier began thinking about implementing gradient-based MCMC samplers, and developed the mcex package to experiment with his ideas. The following year, John was invited by the team to re-engineer PyMC to accomodate Hamiltonian Monte Carlo sampling. This led to the adoption of Theano as the computational back end, and marked the beginning of PyMC3’s development. The first alpha version of PyMC3 was released in June 2015. Over the following 2 years, the core development team grew to 12 members, and the first release, PyMC3 3.0, was launched in January 2017.
Support PyMC¶
PyMC3 is a non-profit project under NumFOCUS umbrella. If you want to support PyMC3 financially, consider donating to the project.
Citing PyMC3¶
If you use PyMC3 in your reseach please cite: Salvatier J., Wiecki T.V., Fonnesbeck C. (2016) Probabilistic programming in Python using PyMC3. PeerJ Computer Science 2:e55 [DOI: 10.7717/peerj-cs.55](https://doi.org/10.7717/peerj-cs.55).
The BibTeX entry is:
@article{pymc3,
title={Probabilistic programming in Python using PyMC3},
author={Salvatier, John and Wiecki, Thomas V and Fonnesbeck, Christopher},
journal={PeerJ Computer Science},
volume={2},
pages={e55},
year={2016},
publisher={PeerJ Inc.}
}
PyMC for enterprise¶
PyMC is now available as part of the Tidelift Subscription!
Tidelift is working with PyMC and the maintainers of thousands of other open source projects to deliver commercial support and maintenance for the open source dependencies you use to build your applications. Save time, reduce risk, and improve code health, while contributing financially to PyMC – making it even more robust, reliable and, let’s face it, amazing!
Enterprise-ready open source software — managed for you¶
The Tidelift Subscription is a managed open source subscription for application dependencies covering millions of open source projects across JavaScript, Python, Java, PHP, Ruby, .NET, and more. And now, your favorite probabilistic programming language is included in the Tidelift subscription!
Your subscription includes:
Security updates: Tidelift’s security response team coordinates patches for new breaking security vulnerabilities and alerts immediately through a private channel, so your software supply chain is always secure.
Licensing verification and indemnification: Tidelift verifies license information to enable easy policy enforcement and adds intellectual property indemnification to cover creators and users in case something goes wrong. You always have a 100% up-to-date bill of materials for your dependencies to share with your legal team, customers, or partners.
Maintenance and code improvement: Tidelift ensures the software you rely on keeps working as long as you need it to work. Your managed dependencies are actively maintained and Tidelift recruits additional maintainers where required.
Package selection and version guidance: Tidelift helps you choose the best open source packages from the start—and then guides you through updates to stay on the best releases as new issues arise.
Roadmap input: Take a seat at the table with the creators behind the software you use. PyMC developers and other Tidelift’s participating maintainers earn more income as our software is used by more subscribers, so we’re interested in knowing what you need.
Tooling and cloud integration: Tidelift works with GitHub, GitLab, BitBucket, and more. It supports every cloud platform (and other deployment targets, too).
The end result? All of the capabilities you expect from commercial-grade software, for the full breadth of open source you use. That means less time grappling with esoteric open source trivia, and more time building your own applications — and your business.
Sponsors¶
More details about sponsoring PyMC3 can be found here. If you are interested in becoming a sponsor, reach out to pymc.devs@gmail.com
Usage Overview¶
For a detailed overview of building models in PyMC3, please read the appropriate sections in the rest of the documentation. For a flavor of what PyMC3 models look like, here is a quick example.
First, let’s import PyMC3 and ArviZ (which handles plotting and diagnostics):
import arviz as az
import numpy as np
import pymc3 as pm
Models are defined using a context manager (with statement). The model is specified declaratively inside the context manager, instantiating model variables and transforming them as necessary. Here is an example of a model for a bioassay experiment:
# Set style
az.style.use("arviz-darkgrid")
# Data
n = np.ones(4)*5
y = np.array([0, 1, 3, 5])
dose = np.array([-.86,-.3,-.05,.73])
with pm.Model() as bioassay_model:
# Prior distributions for latent variables
alpha = pm.Normal('alpha', 0, sigma=10)
beta = pm.Normal('beta', 0, sigma=1)
# Linear combination of parameters
theta = pm.invlogit(alpha + beta * dose)
# Model likelihood
deaths = pm.Binomial('deaths', n=n, p=theta, observed=y)
Save this file, then from a python shell (or another file in the same directory), call:
with bioassay_model:
# Draw samples
trace = pm.sample(1000, tune=2000, cores=2)
# Plot two parameters
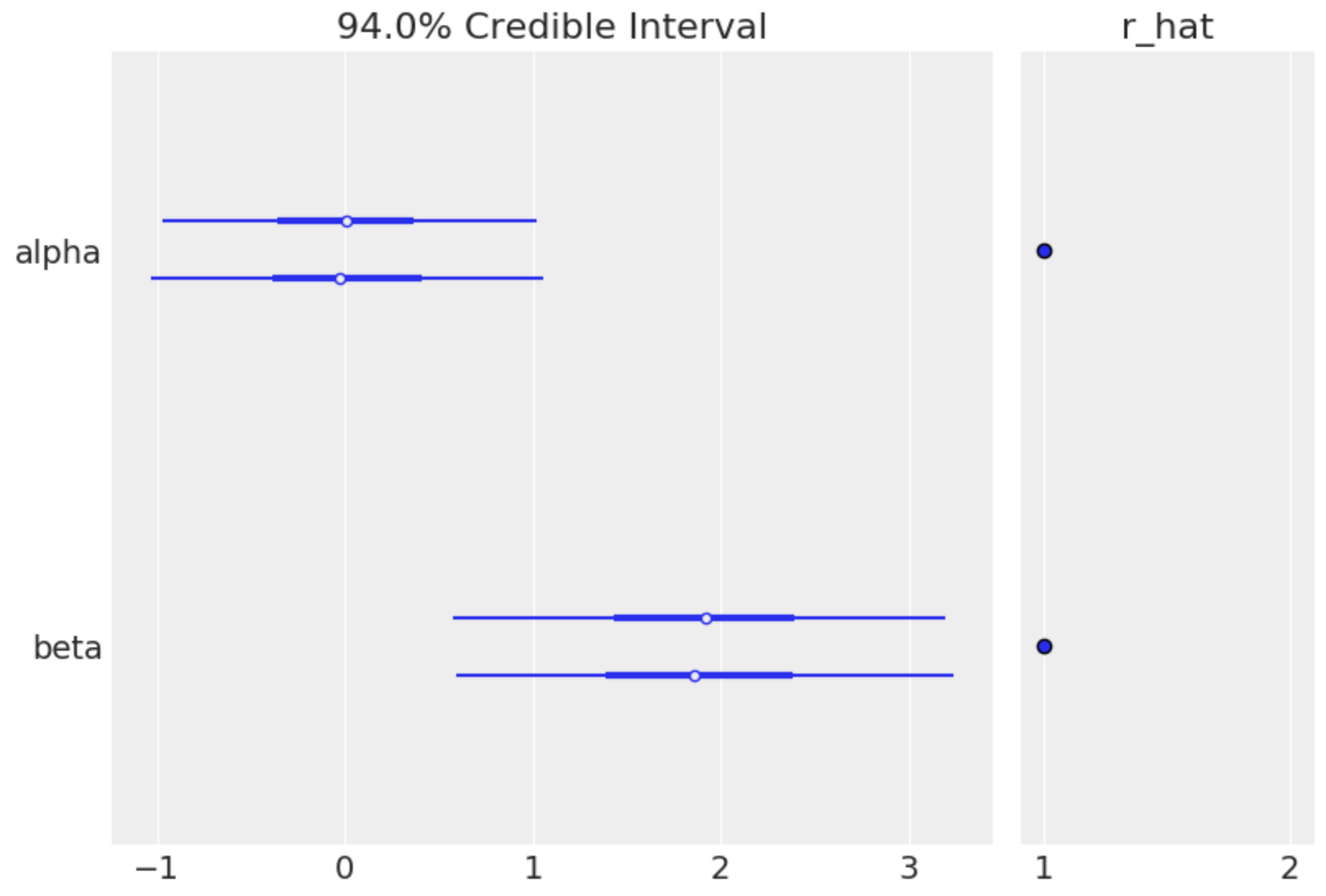
az.plot_forest(trace, var_names=['alpha', 'beta'], r_hat=True)
This example will generate 1000 posterior samples on each of two cores using the NUTS algorithm, preceded by 2000 tuning samples (these are good default numbers for most models).
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [beta, alpha]
|██████████████████████████████████████| 100.00% [6000/6000 00:04<00:00 Sampling 2 chains, 0 divergences]
The sample is returned as arrays inside a MultiTrace object, which is then passed to the plotting function. The resulting graph shows a forest plot of the random variables in the model, along with a convergence diagnostic (R-hat) that indicates our model has converged.